Precision Cancer Screenings
Research center
Understand the science behind the results by reading the information below. Have questions? Contact us today!
The studies and research behind the effectiveness of our tests
Drugs and alcohol have noticeable effects on the immune system’s white blood cells, which protect the body against infectious disease and foreign invaders. Changes to these cells are caused by the direct toxic effects of chemicals (smoking and alcohol). The larger the amount and longer the duration of substance use, the greater the amount of change that occurs.
To counteract these chemical effects, white blood cells “reprogram” their genome (complete DNA set) so they can produce enzymes that metabolize toxins and inflammatory factors. Along the way, they signal other cells to help in their efforts. As they reprogram, the cells don’t change their DNA sequence. Instead, they alter a gene’s methylation (CH3) status, or turn key portions of genes “off” or “on.” Known as “promoters,” this reprogramming controls the amount of enzymes and inflammatory factors produced.
The cell’s alterations of a gene’s methylation status are closely linked to addiction, diseases, and other health changes.
What is the science behind these patented tests, which can detect subtle and not-so-subtle effects of smoking and alcohol on the metabolism of white blood cells? It rests on a solid core of research findings funded by the National Institutes of Health and reproduced by dozens of independent laboratories.
Our state-of-the-art tests detect this stable epigenetic reprogramming by measuring DNA methylation changes in key genes of critical metabolic and inflammatory pathways. These tests focus on the most critical portions of the genome: the promoters. In general, higher levels of DNA methylation are found in inactive genes; active genes tend to have lower DNA methylation levels.
Know your health better than ever before.
No close competitors or no competitor at all.
Cannot be fooled by short-term abstinence.
Compatible with telehealth platforms.
A New Method for Quantifying Smoking

Figure 1. Smoking alters the methylation (CH3) status or “on/off” status of genes. This change in the “on/off” status is closely linked to addiction and other diseases. Our patented assays focus on the most critical portions of the genome – the promoters – genes closely associated with key health outcomes.
Detect Red and Yellow Fluorescent Reporter
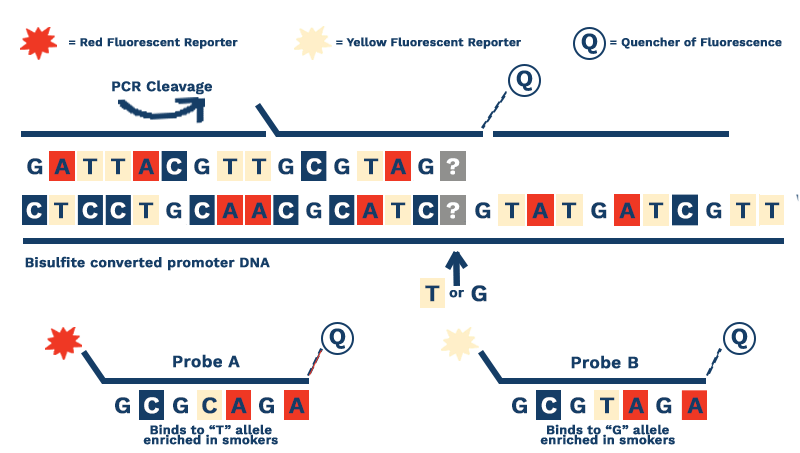
Figure 2. Our assay focuses on the bisulfite converted DNA from the promoters of key genes. Bisulfite conversion, a process invented in the 1990’s, changes non-methylated cytosine bases to uracils. Because DNA polymerases treat uracil like thymine, we can use probes that contain adenosine (A) or cytosine (C) to determine the percent methylation at a given methylation site.
Bibliography
PHILIBERT
Miller S, A Mills J, Long J, Philibert R. A Comparison of the Predictive Power of DNA Methylation with Carbohydrate Deficient Transferrin for Heavy Alcohol Consumption. Epigenetics. 2020 Oct 26:1-11.https://doi.org/10.
Philibert R, Miller S, Noel A, et al. A Four Marker Digital PCR Toolkit for Detecting Heavy Alcohol Consumption and the Effectiveness of Its Treatment. J Insur Med. 2019;48(1):90-102. doi:10.17849/insm-48-1-1-1.1
Genome Wide and Digital PCR Epigenetic Assessments of Alcohol Consumption.
Philibert R, Dogan M, Noel A, et al. Genome-wide and digital polymerase chain reaction epigenetic assessments of alcohol consumption. Am J Med Genet B Neuropsychiatr Genet. 2018;177(5):479-488. doi:10.1002/ajmg.b.32636
A Review of Epigenetic Markers of Tobacco and Alcohol Consumption.
Philibert R, Erwin C. A Review of Epigenetic Markers of Tobacco and Alcohol Consumption. Behav Sci Law. 2015;33(5):675-690. doi:10.1002/bsl.2202
Philibert RA, Penaluna B, White T, et al. A pilot examination of the genome-wide DNA methylation signatures of subjects entering and exiting short-term alcohol dependence treatment programs. Epigenetics. 2014;9(9):1212-1219. doi:10.4161/epi.32252
Cited over 100 times
The impact of recent alcohol use on genome wide DNA methylation signatures.
Philibert RA, Plume JM, Gibbons FX, Brody GH and Beach SRH (2012) The impact of recent alcohol use on genome wide DNA methylation signatures. Front. Gene. 3:54. doi: 10.3389/fgene.2012.00054
AFFILIATED
Current and Future Prospects for Epigenetic Biomarkers of Substance Use Disorders.
Andersen AM, Dogan MV, Beach SR, Philibert RA. Current and Future Prospects for Epigenetic Biomarkers of Substance Use Disorders. Genes (Basel). 2015;6(4):991‐1022. Published 2015 Oct 14. doi:10.3390/genes604099
INDEPENDENT
Brückmann C, Islam SA, MacIsaac JL, et al. DNA methylation signatures of chronic alcohol dependence in purified CD3+ T-cells of patients undergoing alcohol treatment. Sci Rep. 2017;7(1):6605. Published 2017 Jul 26. doi:10.1038/s41598-017-06847-z
Brückmann C, Di Santo A, Karle KN, Batra A, Nieratschker V. Validation of differential GDAP1 DNA methylation in alcohol dependence and its potential function as a biomarker for disease severity and therapy outcome. Epigenetics. 2016;11(6):456-463. doi:10.1080/15592294.2016.1179411
PHILIBERT
DNA methylation differentiates smoking from vaping and non-combustible tobacco use.
Andersen A, Reimer R, Dawes K, Becker A, Hutchens N, Miller S, Dogan M, Hundley B, A Mills J, D Long J, Philibert R. DNA methylation differentiates smoking from vaping and non-combustible tobacco use. Epigenetics. 2021 Feb 25:1-13. https://doi.org/10.
Array-Based Epigenetic Aging Indices May Be Racially Biased.
Philibert R, Beach SRH, Lei M-K, Gibbons FX, Gerrard M, Simons RL, Dogan MV. Array-Based Epigenetic Aging Indices May Be Racially Biased. Genes. 2020; 11(6):685. https://doi.org/10.3390/
Philibert R, Mills JA, Long JD, Salisbury SE, Comellas A, Gerke A, Dawes K, Vander Weg M, Hoffman EA. The Reversion of cg05575921 Methylation in Smoking Cessation: A Potential Tool for Incentivizing Healthy Aging. Genes. 2020; 11(12):1415. https://doi.org/10.3390/
AHRR methylation predicts smoking status and smoking intensity in both saliva and blood DNA.
Philibert R, Dogan M, Beach SRH, Mills JA, Long JD. AHRR methylation predicts smoking status and smoking intensity in both saliva and blood DNA. Am J Med Genet B Neuropsychiatr Genet. 2020;183(1):51-60. doi:10.1002/ajmg.b.32760
AHRR Methylation Is a Significant Predictor of Mortality Risk in Framingham Heart Study
Philibert RA, Dogan MV, Mills JA, Long JD. AHRR Methylation is a Significant Predictor of Mortality Risk in Framingham Heart Study. J Insur Med. 2019;48(1):79‐89. doi:10.17849/insm-48-1-1-11.1
Reversion of AHRR Demethylation Is a Quantitative Biomarker of Smoking Cessation.
Philibert R, Hollenbeck N, Andersen E, et al. Reversion of AHRR Demethylation Is a Quantitative Biomarker of Smoking Cessation. Front Psychiatry. 2016;7:55. Published 2016 Apr 6. doi:10.3389/fpsyt.2016.00055
A Quantitative Epigenetic Approach for the Assessment of Cigarette Consumption.
Philibert R, Hollenbeck N, Andersen E, et al. A quantitative epigenetic approach for the assessment of cigarette consumption. Front Psychol. 2015;6:656. Published 2015 Jun 2. doi:10.3389/fpsyg.2015.00656
Coordinated Changes in AHRR Methylation in Lymphoblasts and Pulmonary Macrophages from Smokers.
Monick MM, Beach SR, Plume J, et al. Coordinated changes in AHRR methylation in lymphoblasts and pulmonary macrophages from smokers. Am J Med Genet B Neuropsychiatr Genet. 2012;159B(2):141‐151. doi:10.1002/ajmg.b.32021
Cited over 200 times
Philibert, R.A., Beach, S.R.H., Lei, M. et al. Changes in DNA methylation at the aryl hydrocarbon receptor repressor may be a new biomarker for smoking. Clin Epigenet 5, 19 (2013). https://doi.org/10.1186/1868-7083-5-19
Cited over 100 times
Lei MK, Gibbons FX, Simons RL, Philibert RA, Beach SRH. The Effect of Tobacco Smoking Differs across Indices of DNA Methylation-Based Aging in an African American Sample: DNA Methylation-Based Indices of Smoking Capture These Effects. Genes (Basel). 2020;11(3):311. Published 2020 Mar 14. doi:10.3390/genes11030311
Andersen AM, Lei MK, Beach SRH, Philibert RA, Sinha S, Colgan JD. Cigarette and Cannabis Smoking Effects on GPR15+ Helper T Cell Levels in Peripheral Blood: Relationships with Epigenetic Biomarkers. Genes (Basel). 2020;11(2):149. Published 2020 Jan 30. doi:10.3390/genes11020149
AHRR methylation predicts smoking status and smoking intensity in both saliva and blood DNA.
Philibert R, Dogan M, Beach SRH, Mills JA, Long JD. AHRR methylation predicts smoking status and smoking intensity in both saliva and blood DNA. Am J Med Genet B Neuropsychiatr Genet. 2020;183(1):51-60. doi:10.1002/ajmg.b.32760
Saliva DNA Methylation Detects Nascent Smoking in Adolescents.
Dawes K, Andersen A, Vercande K, et al. Saliva DNA Methylation Detects Nascent Smoking in Adolescents. J Child Adolesc Psychopharmacol. 2019;29(7):535-544. doi:10.1089/cap.2018.0176
Philibert R, Dogan M, Noel A, Miller S, Krukow B, Papworth E, Cowley J, Long JD, Beach SRH and Black DW (2018) Dose Response and Prediction Characteristics of a Methylation Sensitive Digital PCR Assay for Cigarette Consumption in Adults. Front. Genet. 9:137.
Andersen AM, Philibert RA, Gibbons FX, Simons RL, Long J. Accuracy and utility of an epigenetic biomarker for smoking in populations with varying rates of false self-report. Am J Med Genet B Neuropsychiatr Genet. 2017;174(6):641-650. doi:10.1002/ajmg.b.32555
A Quantitative Epigenetic Approach for the Assessment of Cigarette Consumption.
Philibert R, Hollenbeck N, Andersen E, et al. A quantitative epigenetic approach for the assessment of cigarette consumption. Front Psychol. 2015;6:656. Published 2015 Jun 2. doi:10.3389/fpsyg.2015.00656
Dogan MV, Shields B, Cutrona C, et al. The effect of smoking on DNA methylation of peripheral blood mononuclear cells from African American women. BMC Genomics. 2014;15:151. Published 2014 Feb 22. doi:10.1186/1471-2164-15-151
Philibert RA, Beach SR, Lei MK, Brody GH. Changes in DNA methylation at the aryl hydrocarbon receptor repressor may be a new biomarker for smoking. Clin Epigenetics. 2013;5(1):19. Published 2013 Oct 11. doi:10.1186/1868-7083-5-19
Demethylation of the Aryl Hydrocarbon Receptor Repressor as a Biomarker for Nascent Smokers.
Philibert RA, Beach SR, Brody GH. Demethylation of the aryl hydrocarbon receptor repressor as a biomarker for nascent smokers. Epigenetics. 2012;7(11):1331-1338. doi:10.4161/epi.22520
Philibert RA, Beach SR, Brody GH. Demethylation of the aryl hydrocarbon receptor repressor as a biomarker for nascent smokers. Epigenetics. 2012;7(11):1331-1338. doi:10.4161/epi.22520
AFFILIATED
Lei MK, Gibbons FX, Simons RL, Philibert RA, Beach SRH. The Effect of Tobacco Smoking Differs across Indices of DNA Methylation-Based Aging in an African American Sample: DNA Methylation-Based Indices of Smoking Capture These Effects. Genes (Basel). 2020;11(3):311. Published 2020 Mar 14. doi:10.3390/genes11030311
Saliva DNA Methylation Detects Nascent Smoking in Adolescents.
Dawes K, Andersen A, Vercande K, Papworth E, Philibert W, Beach SRH, Gibbons FX, Gerrard M, Philibert R.
J Child Adolesc Psychopharmacol. 2019 Aug;29(7):535-544. doi: 10.1089/cap.2018.0176. Epub 2019 Jun 10.
A Droplet Digital PCR Assay for Smoking Predicts All-Cause Mortality.
Andersen AM, Ryan PT, Gibbons FX, Simons RL, Long JD, Philibert RA. A Droplet Digital PCR Assay for Smoking Predicts All-Cause Mortality. J Insur Med. 2018;47(4):220-229. doi:10.17849/insm-47-4-1-10.1
INDEPENDENT
Validated Inference of Smoking Habits From Blood With a Finite DNA Methylation Marker Set
Maas SCE, Vidaki A, Wilson R, et al. Validated inference of smoking habits from blood with a finite DNA methylation marker set. Eur J Epidemiol. 2019;34(11):1055‐1074. doi:10.1007/s10654-019-00555-w
AHRR (cg05575921) Hypomethylation Marks Smoking Behaviour, Morbidity and Mortality
Bojesen SE, Timpson N, Relton C, Davey Smith G, Nordestgaard BG. AHRR (cg05575921) hypomethylation marks smoking behaviour, morbidity and mortality. Thorax. 2017;72(7):646‐653. doi:10.1136/thoraxjnl-2016-208789
Zhang Y, Schöttker B, Florath I, et al. Smoking-Associated DNA Methylation Biomarkers and Their Predictive Value for All-Cause and Cardiovascular Mortality. Environ Health Perspect. 2016;124(1):67‐74. doi:10.1289/ehp.1409020
Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin Epigenetics. 2015;7:113. Published 2015 Oct 16. doi:10.1186/s13148-015-0148-3
DNA Methylation as a Long-Term Biomarker of Exposure to Tobacco Smoke.
Shenker NS, Ueland PM, Polidoro S, et al. DNA methylation as a long-term biomarker of exposure to tobacco smoke. Epidemiology. 2013;24(5):712-716. doi:10.1097/EDE.0b013e31829d5cb3
Shenker NS, Polidoro S, van Veldhoven K, et al. Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Hum Mol Genet. 2013;22(5):843-851. doi:10.1093/hmg/dds488
Joubert BR, Håberg SE, Nilsen RM, et al. 450K epigenome-wide scan identifies differential DNA methylation in newborns related to maternal smoking during pregnancy [published correction appears in Environ Health Perspect. 2012 Dec;120(12):A455]. Environ Health Perspect. 2012;120(10):1425-1431. doi:10.1289/ehp.1205412
PHILIBERT
A simple, rapid, interpretable, actionable and implementable digital PCR based mortality index.
Philibert R, Long JD, Mills JA, Beach SRH, Gibbons FX, Gerrard M, Simons R, Pinho PB, Ingle D, Dawes K, Dogan T, Dogan M. A simple, rapid, interpretable, actionable and implementable digital PCR based mortality index. Epigenetics. 2020 Nov 2:1-15. https://doi.org/10.
AHRR Methylation is a Significant Predictor of Mortality Risk in Framingham Heart Study.
Philibert RA, Dogan MV, Mills JA, Long JD. AHRR Methylation is a Significant Predictor of Mortality Risk in Framingham Heart Study. J Insur Med. 2019;48(1):79-89. doi:10.17849/insm-48-1-1-11.1
Mills JA, Beach SRH, Dogan M, et al. A Direct Comparison of the Relationship of Epigenetic Aging and Epigenetic Substance Consumption Markers to Mortality in the Framingham Heart Study. Genes (Basel). 2019;10(1):51. Published 2019 Jan 15. doi:10.3390/genes10010051
A Droplet Digital PCR Assay for Smoking Predicts All-Cause Mortality
Andersen AM, Ryan PT, Gibbons FX, Simons RL, Long JD, Philibert RA. A Droplet Digital PCR Assay for Smoking Predicts All-Cause Mortality. J Insur Med. 2018;47(4):220-229. doi:10.17849/insm-47-4-1-10.1
